|
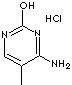
5-METHYLCYTOSINE HCl |
| 4-Amino-5-methyl-2(1H)-pyrimidinone HCl; 4-Amino-5-methyl-1H-pyrimidin-2-one hydrochloride; 4-Amino-5-methyl-3H-pyrimidin-2-one hydrochloride; 4-Amino-2-hydroxy-5-methylpyrimidine; |
|
|
| PRODUCT IDENTIFICATION |
|
|
CAS RN |
58366-64-6, 554-01-8 (parent) |
|
EINECS RN |
261-223-9, 209-058-3 (parent) |
|
FORMULA |
C5H7N3O·HCl |
|
MOLE WEIGHT |
161.59 |
|
H.S CODE |
2933.59.7000 |
|
SMILES |
n1c(c(C)cnc1O)N.Cl |
|
CLASSIFICATION |
Nucleoside Analog |
|
EXTRA NOTES |
|
|
|
| PHYSICAL AND CHEMICAL PROPERTIES |
|
|
PHYSICAL STATE |
white to off-white powder |
|
MELTING POINT |
298 C |
|
BOILING POINT |
|
|
DENSITY |
|
|
SOLUBILITY IN WATER |
|
|
pH |
|
|
VAPOR DENSITY |
|
|
REFRACTIVE INDEX |
|
|
FLASH POINT |
|
|
|
| STABILITY AND REACTIVITY | |
| STABILITY | Stable under normal conditions. |
|
INCOMPATIBLE MATERIALS |
Strong oxidizing agents |
| DECOMPOSITION PRODUCTS |
Carbon oxides |
| POLYMERIZATION | Has not been reported |
|
NFPA RATINGS |
Health Hazard: 1, Fire: 0, Reactivity Hazard: 0 |
|
|
| SAFETY INFORMATION |
|
|
HAZARD NOTES |
|
|
EYE |
May cause eye irritation |
|
SKIN |
May cause skin irritation. May be harmful if absorbed through skin. |
|
INGESTION |
May be harmful if swallowed. |
|
INHALATION |
May be harmful if inhaled. May cause respiratory tract irritation. |
|
CHRONIC |
|
|
|
| TRANSPORT & REGULATORY INFORMATION |
|
|
UN NO. |
Not regulated |
| HAZARD CLASS |
|
| PACKING GROUP |
|
| HAZARD SYMBOL |
|
|
RISK PHRASES |
36/37/38: Irritating to eyes, respiratory system and skin |
|
SAFETY PHRASES |
24/25: Avoid contact with skin and eyes. |
|
|
| EXTERNAL LINKS & GENERAL DESCRIPTION |
|
|
Chemical modification of DNA is one way that cells regulate which genes are
expressed as proteins. Methylation of certain cytosines is a chemical
modification that regulates gene expression in many normal biological contexts.
In addition, inactivation of tumor suppressor genes by aberrant methylation of
cytosines is frequently associated with cancer.
(http://innovation.swmed.edu/)
As a result of the above observations, universal bases have been used to increase the effective size of short primers without increasing multiplicity. A number of analogues were tested to improve the effectiveness of octamers for cycle sequencing reactions. Of the analogues tested, 11 proved to be most effective. Normal octamers are at best poor primers in cycle sequencing, requiring low reaction temperatures. The use of up to four 11 residues at the 5'-end of the primer resulted in much improved performance. The use of other modified bases, such as 5-methylcytosine and diaminopurine (to replace cytosine and adenine), in addition to a tail of 11, resulted in 8mers, now effectively 12mers, that were effective at an extension temperature of 55°C (unpublished data). (http://nar.oxfordjournals.org/) The identification of methylated sites on bacterial genomic DNA would be a useful tool to study the major roles of DNA methylation in prokaryotes: distinction of self and nonself DNA, direction of post-replicative mismatch repair, control of DNA replication and cell cycle, and regulation of gene expression. Three types of methylated nucleobases are known: N6-methyladenine, 5-methylcytosine and N4-methylcytosine. The aim of this study was to develop a method to detect all three types of DNA methylation in complete genomic DNA. It was previously shown that N6-methyladenine and 5-methylcytosine in plasmid and viral DNA can be detected by intersequence trace comparison of methylated and unmethylated DNA. We extended this method to include N4-methylcytosine detection in both in vitro and in vivo methylated DNA. Furthermore, application of intersequence trace comparison was extended to bacterial genomic DNA. Finally, we present evidence that intrasequence comparison suffices to detect methylated sites in genomic DNA. In conclusion, we present a method to detect all three natural types of DNA methylation in bacterial genomic DNA. This provides the possibility to define the complete methylome of any prokaryote. (http://pt.wkhealth.com/) Unnatural nucleobases have proven to be extremely interesting in terms of their effects on the properties of DNA. The design of novel base pairs is being exploited to add functionality to the nucleic acid and is only limited by the creativity of the chemist. Future work will likely use nucleobase analogues to elucidate the mechanisms of DNA binding proteins, inhibit DNA polymerases in an antineoplastic or anti-viral context, and for the design of novel nanomaterials. (http://organicdivision.org/)
|
|
|
| SALES SPECIFICATION |
|
|
APPEARANCE |
white to off-white powder |
|
ASSAY |
99.0% min |
|
|
| PACKING |
|
|
|
|
| PRICE INFORMATION |
|
|